normerr
Returns the normalized error squared for signals in x with
covariance C. For multiple runs, returns the n-run average normalized
error squared. Returns also the fraction of points within the 100% * p
probability bounds based on the number of runs. This can be used to
determine if errors in x correspond appropriately to the covariance.
[err, f, b, errs] = normerr(x, C)
[err, f, b, errs] = normerr(x, C, p)
[err, f, b, errs] = normerr(x, C, p, type)
[err, f, b, errs] = normerr(x, C, p, type, (etc.))Inputs
x | Residuals. For estimation error, |
|---|---|
C | Covariance stored as |
p | Probability region (0 to 1) to examine. E.g., to compare the normalized residuals with a 95% probability region, use 0.95. Default is 0.95. |
type | For type 1, the error |
(etc.) | Option-value pairs: |
Outputs
err | N-run average normalized error (1-by- |
|---|---|
f | Fraction of points in |
b | Probability bounds stored as |
errs | Matrix containing normalized error for each run individually
(1-by- |
Example
Generate a large number of runs and samples per run of a 3-dimensional
state with a different covariance matrix everywhere. See that the
theoretical bounds and the empirical bounds determined by normerr match
well.
% Define the sizes and preallocate space for the errors and covariance
% matrices.
nx = 3; % 3 states
nr = 25; % 25 runs
ns = 1000; % 1000 samples each
x = zeros(nx, ns, nr); % A place for all the residual data
C = zeros(nx, nx, ns, nr); % A place for each covariance matrix
% For each sample of each run, generate a random positive-definite
% covariance matrix and make a random draw from it.
for r = 1:nr
for s = 1:ns
C(:, :, s, r) = randcov(nx);
x(:, s, r) = mnddraw(C(:, :, s, r), 1);
end
endExamine what fraction of the data lay within the 90% confidence bounds (should be close to 0.90).
% For type 1 -- total state error
figure(1);
subplot(2, 1, 1);
[err1, f1] = normerr(x, C, 0.90, 1, 'Plot', true);
% For type 2 -- individual state error
subplot(2, 1, 2);
[err2, f2] = normerr(x, C, 0.90, 2, 'Plot', true);Percent of data in theoretical 90.0% bounds: 88.1%
Percent of data in theoretical 90.0% bounds: 89.6%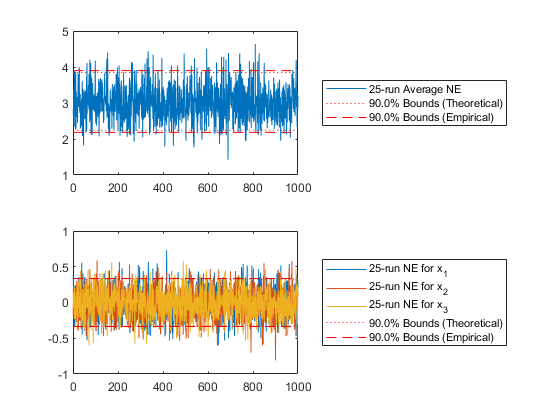
See Also
*kf v1.0.3 January 17th, 2025
©2025 An Uncommon Lab